Data visualization with ggplot2
ggplot
ggplot2 is a tidyverse package for data visualization. It follows the "grammar of graphics"
ggplot
ggplot2 is a tidyverse package for data visualization. It follows the "grammar of graphics"
Even with just a few of its functions, it can make beautiful, publication-ready plots.
ggplot
ggplot2 is a tidyverse package for data visualization. It follows the "grammar of graphics"
Even with just a few of its functions, it can make beautiful, publication-ready plots.
Its more advanced features add even more versatility.
ggplot
ggplot2 is a tidyverse package for data visualization. It follows the "grammar of graphics"
Even with just a few of its functions, it can make beautiful, publication-ready plots.
Its more advanced features add even more versatility.
People make companion packages that allow you to do even more, e.g., ggpubr to add statistics to your plot -- you'll use this in gradstats.
Palmer penguins
We're going to use the Palmer Penguins dataset as an example throughout our discussion of ggplot.
Palmer penguins
We're going to use the Palmer Penguins dataset as an example throughout our discussion of ggplot.
This data comes from the palmerpenugins package, which you can download from CRAN using install.packages("palmerpenguins").
Palmer penguins
We're going to use the Palmer Penguins dataset as an example throughout our discussion of ggplot.
This data comes from the palmerpenugins package, which you can download from CRAN using install.packages("palmerpenguins").
And remember, to load the package, use library(penguins), which will give you access to the built-in dataset, called penguins
Palmer penguins
We're going to use the Palmer Penguins dataset as an example throughout our discussion of ggplot.
This data comes from the palmerpenugins package, which you can download from CRAN using install.packages("palmerpenguins").
And remember, to load the package, use library(penguins), which will give you access to the built-in dataset, called penguins

Palmer penguins
library(palmerpenguins)glimpse(penguins)## Rows: 344## Columns: 8## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adeli…## $ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgersen, Torgersen, Torgersen, …## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, 42.0, 37.8, 37.8, 41.1, 3…## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, 20.2, 17.1, 17.3, 17.6, 2…## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186, 180, 182, 191, 198, 185…## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, 4250, 3300, 3700, 3200, 3…## $ sex <fct> male, female, female, NA, female, male, female, male, NA, NA, NA, NA, female,…## $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007,…ggplot2
When creating a plot in ggplot2, the first thing you have to do is call the ggplot() function.
ggplot2
When creating a plot in ggplot2, the first thing you have to do is call the ggplot() function.
It takes data as its first argument. Let's start there...
ggplot2
All we got was a blank square!
ggplot2
All we got was a blank square!
To understand why, let's back up to the philosophy of ggplot...
ggplot2
What is a statistical graphic?
ggplot2
What is a statistical graphic?
According to Hadley Wickham, we can think of statistical graphics as:
ggplot2
What is a statistical graphic?
According to Hadley Wickham, we can think of statistical graphics as:
- Mappings from data to aesthetic attributes (color, shape, size)
- Consisting of geometric objects (points, lines, bars)
- On a particular coordinate system (or scale)
- Sometimes derived from a statistical transformation
ggplot2
What is a statistical graphic?
According to Hadley Wickham, we can think of statistical graphics as:
- Mappings from data to aesthetic attributes (color, shape, size)
- Consisting of geometric objects (points, lines, bars)
- On a particular coordinate system (or scale)
- Sometimes derived from a statistical transformation
In ggplot, that combination of things forms a layer. The ggplot terminology is:
ggplot2
What is a statistical graphic?
According to Hadley Wickham, we can think of statistical graphics as:
- Mappings from data to aesthetic attributes (color, shape, size)
- Consisting of geometric objects (points, lines, bars)
- On a particular coordinate system (or scale)
- Sometimes derived from a statistical transformation
In ggplot, that combination of things forms a layer. The ggplot terminology is:
- aes - aesthetic mappings
- geom - geometric objects
- scale - scales
- stat - statistical transformations
ggplot
Putting all that together, the flow of information in ggplot is...
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...transformed by
statbefore being passed to ageom()...
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...transformed by
statbefore being passed to ageom()...which is placed, sized, and colored according to its relevant
scale...
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...transformed by
statbefore being passed to ageom()...which is placed, sized, and colored according to its relevant
scale...then ta-da! rendered plot.
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...transformed by
statbefore being passed to ageom()...which is placed, sized, and colored according to its relevant
scale...then ta-da! rendered plot.
We already did step 1 above, by providing our penguins dataset to ggplot().
ggplot
Putting all that together, the flow of information in ggplot is...
datais attached to theggplot()call...mapped by
aes()...transformed by
statbefore being passed to ageom()...which is placed, sized, and colored according to its relevant
scale...then ta-da! rendered plot.
We already did step 1 above, by providing our penguins dataset to ggplot().
Now let's move onto aesthetic mapping.
But first... pipes!
%>%
Pipe operator
Pipe operator
Pipes are a way to write code in a way that is easier to read and understand. They allow you to string a series of functions together into a pipeline.
Pipe operator
Pipes are a way to write code in a way that is easier to read and understand. They allow you to string a series of functions together into a pipeline.
%>%
Strictly speaking, a pipe passes an object on the left-hand side as the first argument of whatever function is on the right-hand side.
Pipe operator
Pipes are a way to write code in a way that is easier to read and understand. They allow you to string a series of functions together into a pipeline.
%>%
Strictly speaking, a pipe passes an object on the left-hand side as the first argument of whatever function is on the right-hand side.
In general form, this means that...
x %>% f(y) is the same as f(x, y)
Pipe operator
Pipes are a way to write code in a way that is easier to read and understand. They allow you to string a series of functions together into a pipeline.
%>%
Strictly speaking, a pipe passes an object on the left-hand side as the first argument of whatever function is on the right-hand side.
In general form, this means that...
x %>% f(y) is the same as f(x, y)
The first argument of many tidyverse functions is data. This is the case for ggplot(), as we just saw.
Pipe operator
Pipes are a way to write code in a way that is easier to read and understand. They allow you to string a series of functions together into a pipeline.
%>%
Strictly speaking, a pipe passes an object on the left-hand side as the first argument of whatever function is on the right-hand side.
In general form, this means that...
x %>% f(y) is the same as f(x, y)
The first argument of many tidyverse functions is data. This is the case for ggplot(), as we just saw.
tidyverse functions are designed this way to make it easier to use pipes!
Pipe operator
Let's take another look at the code we just wrote:
ggplot(data = penguins)Pipe operator
Let's take another look at the code we just wrote:
ggplot(data = penguins)We can re-write this code by "piping" the first argument, data to the function, ggplot()
penguins %>% ggplot()Pipe operator
Let's take another look at the code we just wrote:
ggplot(data = penguins)We can re-write this code by "piping" the first argument, data to the function, ggplot()
penguins %>% ggplot()You can think of the pipe as representing the phrase "and then..."
Pipe operator
Let's take another look at the code we just wrote:
ggplot(data = penguins)We can re-write this code by "piping" the first argument, data to the function, ggplot()
penguins %>% ggplot()You can think of the pipe as representing the phrase "and then..."
Verbally, we might describe the code we just wrote as "Start with the penguins data, and then create the foundation of a plot"
Pipe operator
Let's take another look at the code we just wrote:
ggplot(data = penguins)We can re-write this code by "piping" the first argument, data to the function, ggplot()
penguins %>% ggplot()You can think of the pipe as representing the phrase "and then..."
Verbally, we might describe the code we just wrote as "Start with the penguins data, and then create the foundation of a plot"
⌨️ You can enter a pipe with the shortcut CTRL+Shift+M for PC or CMD+Shift+M for Mac.
Your Turn 1
02:00
The code below calculates the mean of the variable flipper_length_mm in the penguins dataset.
mean(penguins$flipper_length_mm, na.rm = TRUE)Run the code chunk provided to calculate the mean of
flipper_length_mm.Re-write the code above using the pipe operator,
%>%. Check that the answer you get matches the original code you ran above.
Now back to ggplot...
Aesthetic mapping
The basic idea with aesthetic mapping is that we want to take our data and map it onto physical space using geometric objects, or geoms.
Aesthetic mapping
The basic idea with aesthetic mapping is that we want to take our data and map it onto physical space using geometric objects, or geoms.
For ggplot to place a geom, it needs to know how to map aspects of the data onto the aesthetics (or visual features) of the graph.
Aesthetic mapping
The basic idea with aesthetic mapping is that we want to take our data and map it onto physical space using geometric objects, or geoms.
For ggplot to place a geom, it needs to know how to map aspects of the data onto the aesthetics (or visual features) of the graph.
Each geom() function in ggplot takes a mapping argument.
Aesthetic mapping
The basic idea with aesthetic mapping is that we want to take our data and map it onto physical space using geometric objects, or geoms.
For ggplot to place a geom, it needs to know how to map aspects of the data onto the aesthetics (or visual features) of the graph.
Each geom() function in ggplot takes a mapping argument.
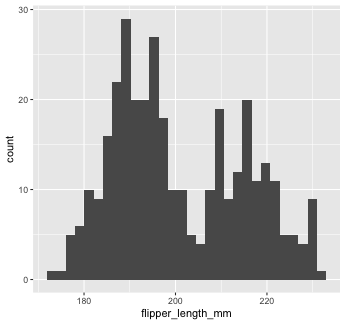
Let’s start with a basic geom, geom_histogram()...
Mapping with geoms
We're going to call ggplot() again, and this time add the geom_histogram() layer, telling it to map the variable flipper_length_mm to the x-axis aesthetic
Mapping with geoms
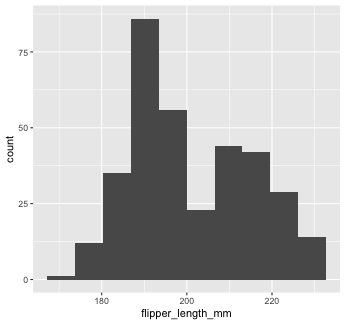
You can add additional arguments to the geom(). For example, we could change the number of bins
Mapping with geoms
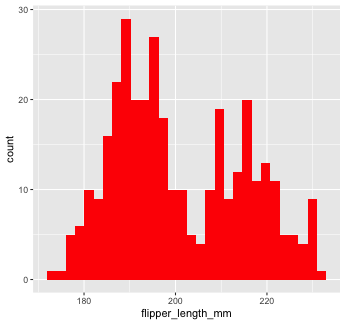
Or make them a different color. We can change the color of 2-d objects with the fill aesthetic
Mapping with geoms
Now we'll use a different geom -- we'll add a layer of points to our plot using geom_point()
penguins %>% ggplot() + geom_point(mapping = aes(x = flipper_length_mm))## Error: geom_point requires the following missing aesthetics: y
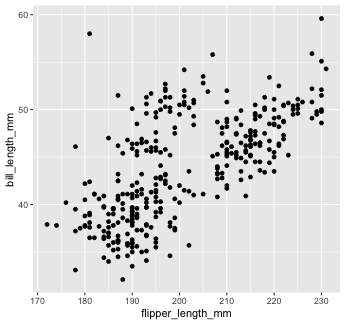
Mapping with geoms
We get an error, telling us that geom_point() requires the y-aesthetic.
This makes sense -- we need an x and y axis to define where points belong on a scatter plot. Let's add bill_length_mm as the y-axis
Mapping with geoms
We get an error, telling us that geom_point() requires the y-aesthetic.
This makes sense -- we need an x and y axis to define where points belong on a scatter plot. Let's add bill_length_mm as the y-axis
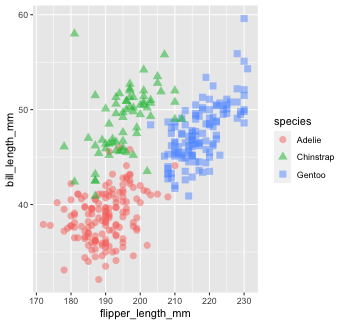
penguins %>% ggplot() + geom_point(mapping = aes(x = flipper_length_mm, y = bill_length_mm))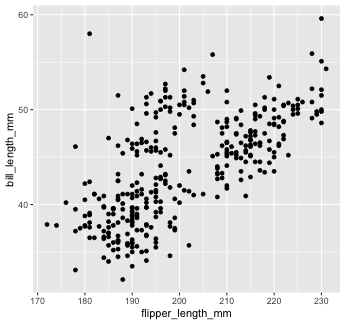
Mapping with geoms
Let's find out if the relationship between flipper_length_mm and bill_length_mm relates to the species of penguin.
We'll map species to the color aesthetic (similar to fill, but for 1-d objects).
Mapping with geoms
Let's find out if the relationship between flipper_length_mm and bill_length_mm relates to the species of penguin.
We'll map species to the color aesthetic (similar to fill, but for 1-d objects).
penguins %>% ggplot() + geom_point( mapping = aes(x = flipper_length_mm, y = bill_length_mm, color = species))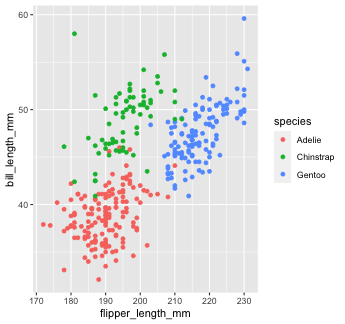
Mapping with geoms
Notice that we included color inside our aesthetic mapping call (mapping = aes()) here, but not when we filled our histogram with the color red earlier
Mapping with geoms
Notice that we included color inside our aesthetic mapping call (mapping = aes()) here, but not when we filled our histogram with the color red earlier
This is the difference between mapping an aesthetic to data and just setting an aesthetic to some value (e.g., "red").
Mapping with geoms
Notice that we included color inside our aesthetic mapping call (mapping = aes()) here, but not when we filled our histogram with the color red earlier
This is the difference between mapping an aesthetic to data and just setting an aesthetic to some value (e.g., "red").
This is a fairly common mistake, so let's take a look at an example
Mapping with geoms
What happens if we tell ggplot to make our points red but accidentally include that inside the aes() call?
Mapping with geoms
What happens if we tell ggplot to make our points red but accidentally include that inside the aes() call?
penguins %>% ggplot() + geom_point(mapping = aes(x = flipper_length_mm, y = bill_length_mm, color = "red"))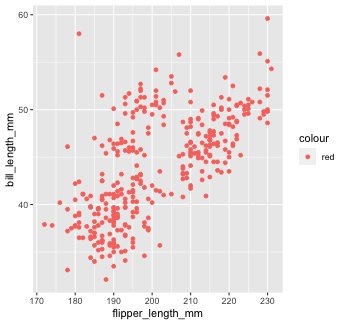
This is not what we want! ggplot is treating the value "red" as if it were part of our data, which it isn't
Your Turn 2
03:30
Create a scatter plot to visualize the relationship between
flipper_length_mmandbill_length_mm.Build on your plot above by adding an aesthetic to visualize the effect of
species. Choose any aesthetic you’d like or play around with a few. What do they do? How might you use more than one aesthetic?
Note: Options for aesthetics include color, shape, size, and alpha (transparency, 0-1 scale).
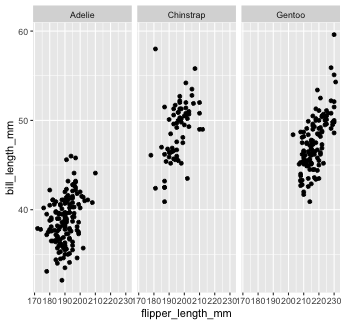
Mapping with geoms
We could also make separate graphs for each species using facet_wrap. We do this by passing a one-sided formula to facet_wrap()
Mapping with geoms
Another thing we often want to do is to add a line over our scatterplot to describe the linear relationship between variables. We can do this by adding a geom_smooth() layer to our plot.
Mapping with geoms
Another thing we often want to do is to add a line over our scatterplot to describe the linear relationship between variables. We can do this by adding a geom_smooth() layer to our plot.
penguins %>% ggplot() + geom_point(aes(x = flipper_length_mm, y = bill_length_mm, color = species)) + geom_smooth(aes(x = flipper_length_mm, y = bill_length_mm), color = "black")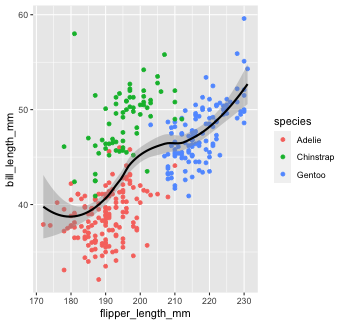
Note that "loess" is the default function for geom_smooth().
Learn more on that here.
You can change that by setting the method argument in geom_smooth(). Let's change it to our old friend linear regression or "lm"
penguins %>% ggplot() + geom_point(aes(x = flipper_length_mm, y = bill_length_mm, color = species)) + geom_smooth(aes(x = flipper_length_mm, y = bill_length_mm), color = "black", method = "lm")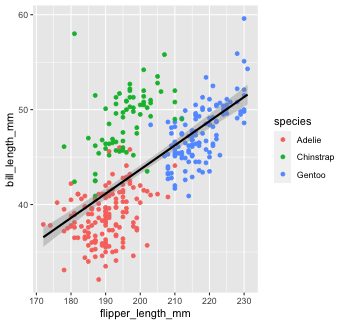
Global aesthetic mapping
Our code so far has been getting rather inefficient. We're specifying the x and y axis for each geom_* call.
Global aesthetic mapping
Our code so far has been getting rather inefficient. We're specifying the x and y axis for each geom_* call.
Instead, we can use global aesthetic mappings, which are specified in the ggplot() call.
Global aesthetic mapping
Our code so far has been getting rather inefficient. We're specifying the x and y axis for each geom_* call.
Instead, we can use global aesthetic mappings, which are specified in the ggplot() call.
Global mappings are inherited by each layer unless they're overwritten.
Global aesthetic mapping
Let's re-make our previous plot using global aesthetic mapping
Global aesthetic mapping
Let's re-make our previous plot using global aesthetic mapping
penguins %>% ggplot(aes(x = flipper_length_mm, y = bill_length_mm))+ geom_point(aes(color = species)) + geom_smooth(color = "black", method = "lm")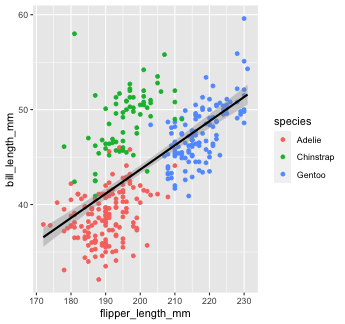
So...what do we put in global aesthetic mapping and what do we put in the aesthetic mapping of specific geoms?
You want to put anything in the global mapping that you want every layer to inherit (or at least the majority of them).
In the code above, I defined the x and y aesthetics globally, because I want those the same in every geom.
However, I don't define thecolor aesthetic globally, because color is geom-specific in this case.
Global aesthetic mapping
Let's take a look at the previous example again, but this time with color in the global aesthetic...
Global aesthetic mapping
Let's take a look at the previous example again, but this time with color in the global aesthetic...
penguins %>% ggplot(aes(x = flipper_length_mm, y = bill_length_mm, color = species))+ geom_point() + # inherit global geom_smooth(method = "lm") #inherit global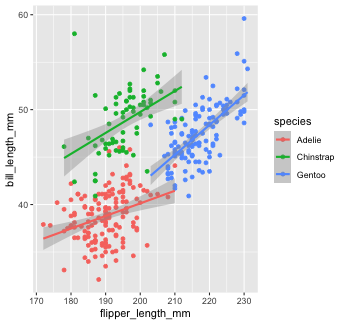
As you can see, global aesthetic mapping gets inherited by every layer. We can override this by providing a different aesthetic mapping in individual geom() calls...
penguins %>% ggplot(aes(x = flipper_length_mm, y = bill_length_mm, color = species))+ geom_point() + #inherit global geom_smooth(method = "lm", color = "black") #override global `color`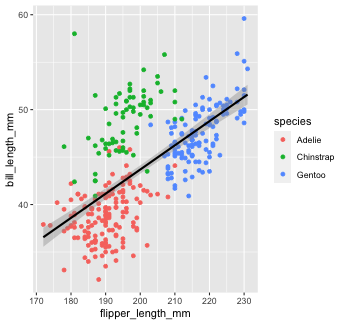
Your Turn 3
04:00
Use
rio::importto read in the datasetpragmatic_scales_data.csv, which is saved inside thedatafolder. Save it to an object calledps_data.Take a glimpse at the data using
glimpse()orstr()to get a sense of the variables. You can alsoView()the data.Fill in the blanks in the code to re-create the plot below. Note: This plot uses a new geom called
geom_col(), which I've filled in for you.
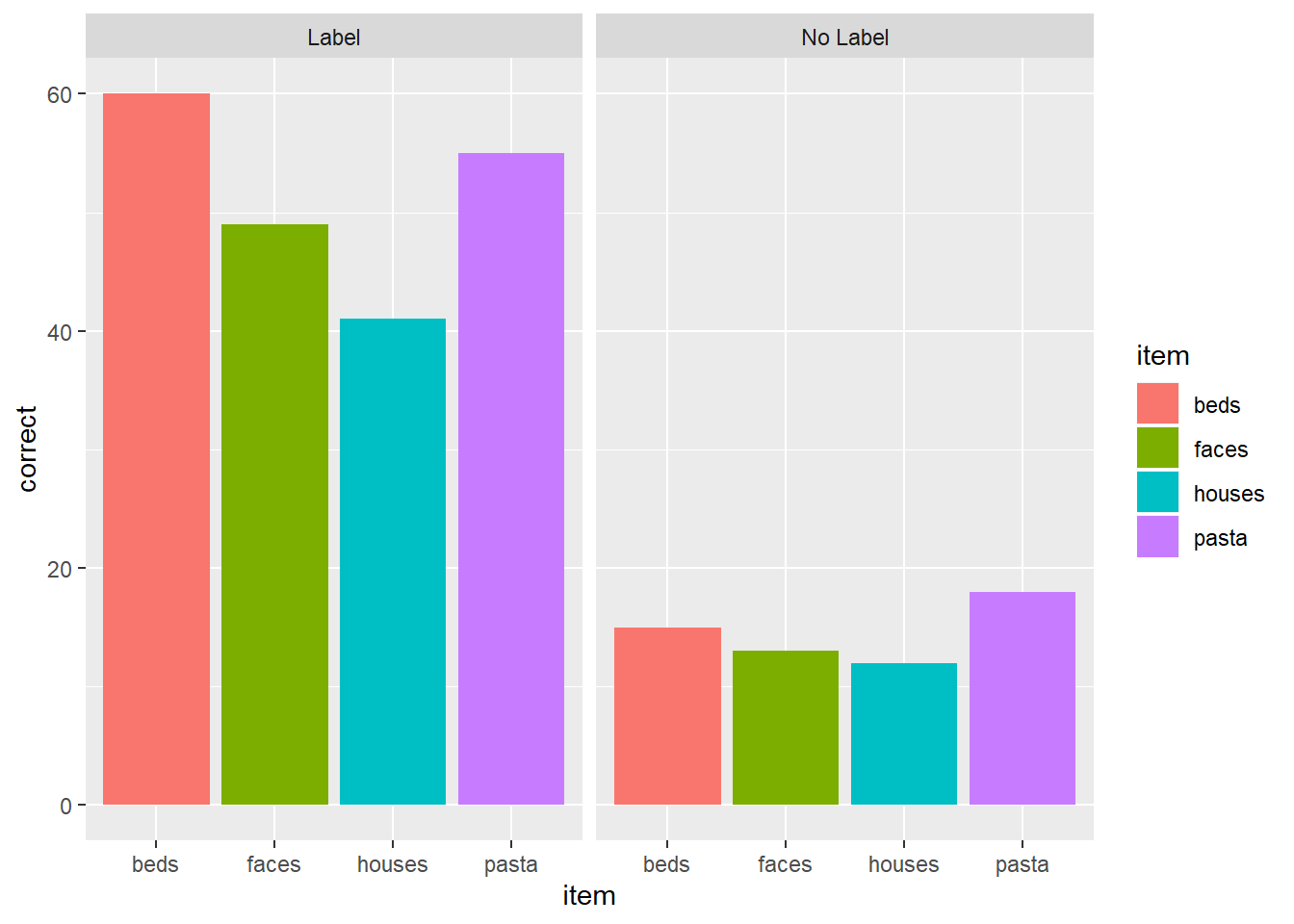
Solution
ps_data <- rio::import(here::here("data/pragmatic_scales_data.csv"))glimpse(ps_data)## Rows: 588## Columns: 5## $ subid <chr> "M22", "M22", "M22", "M22", "T22", "T22", "T22", "T22", "T17", "T17", "T17", "T17", "…## $ item <chr> "faces", "houses", "pasta", "beds", "beds", "faces", "houses", "pasta", "pasta", "fac…## $ correct <int> 1, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1…## $ age <dbl> 2.00, 2.00, 2.00, 2.00, 2.13, 2.13, 2.13, 2.13, 2.32, 2.32, 2.32, 2.32, 2.38, 2.38, 2…## $ condition <chr> "Label", "Label", "Label", "Label", "Label", "Label", "Label", "Label", "Label", "Lab…ps_data %>% ggplot(aes(x = item, y = correct, fill = item)) + geom_col() + facet_wrap(~condition)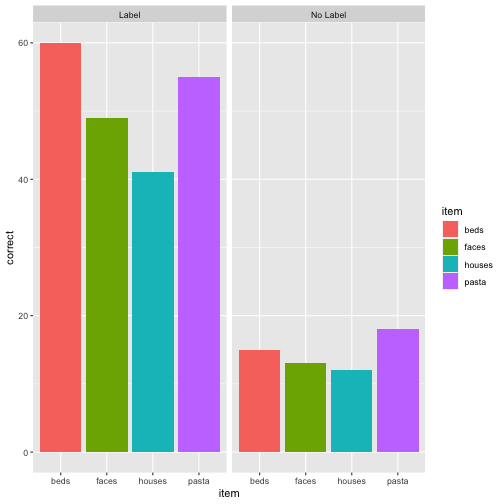
Labels and themes
You can do a TON more customization of your plots than what we've covered so far. The possibilities with ggplot really are endless!
Labels and themes
You can do a TON more customization of your plots than what we've covered so far. The possibilities with ggplot really are endless!
For example, you can change your axis labels, tweak the theme of the plot, etc...
Labels and themes
You can do a TON more customization of your plots than what we've covered so far. The possibilities with ggplot really are endless!
For example, you can change your axis labels, tweak the theme of the plot, etc...
Good resources:
https://ggplot2.tidyverse.org/reference/theme.html
http://r-statistics.co/Complete-Ggplot2-Tutorial-Part2-Customizing-Theme-With-R-Code.html
Let's recap with a flipbook!
Flipbook
Click on the "full screen" icon in the bottom right and use the navigation arrows to flip through the creation of a scatterplot in real time!
Q & A
05:00